Softwares
ChIP-seq

ChromBERT: Uncovering Chromatin State Motifs in the Human Genome Using a BERT-based Approach
[Lee et al, bioRxiv, 2024]

Churros: Epigenome analysis pipeline
[Wang and Nakato, DNA Research, 2023]
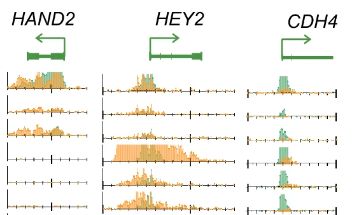
DROMPAplus: Analysis and visualization for multiple ChIP-seq data
[Nakato and Sakata, Methods, 2020]
(This is a successor to DROMPA)
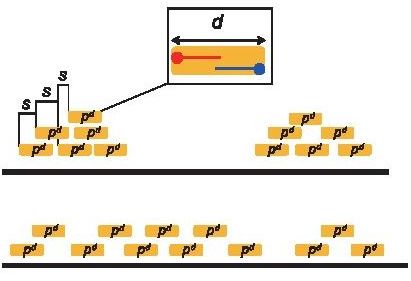
SSP: Sensitive and robust assessment of ChIP-seq read distribution using a strand-shift profile
[Nakato and Shirahige, Bioinformatics, 2018]
RNA-seq

RumBall: Transcriptome analysis pipeline
[Nagai et al., STAR Protocols, 2024]

Clover: Prioritizing rare genes in a differentially expressed gene list
[Oba and Nakato, Genes to Cells, 2024]
Hi-C

CustardPy: 3D genome analysis pipeline
[Nakato et al., Nature Communications, 2023]
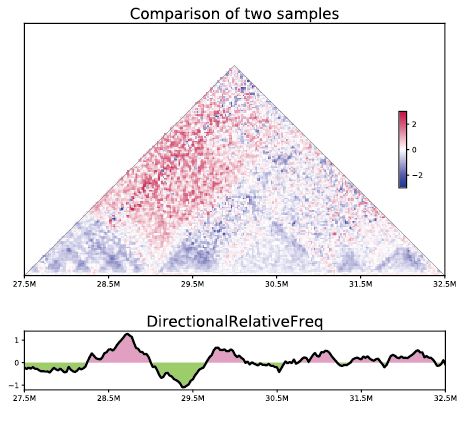
HiC1Dmetrics: Framework to extract various 1D features from Hi-C data
[Wang and Nakato, Briefing in Bioinformatics, 2021]
Single-cell analysis

ShortCake: a Docker-based platform for single-cell analyses
[Nakato and Nagai, Bioinformatics, 2025]
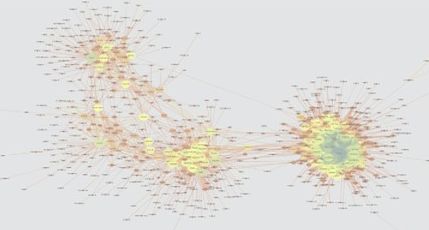
EEISP: Identifying codependent and mutually exclusive gene sets from sparse scRNA-seq data
[Nakajima et al., Nucleic Acids Research, 2021]
Sequence alignment
Cgaln: Fast and space-efficient whole-genome alignment
[Nakato and Gotoh, BMC Bioinformatics, 2010]
Database

CohesinDB: a comprehensive database for cohesin analysis
[Wang and Nakato, Nucleic Acids Research, 2022]
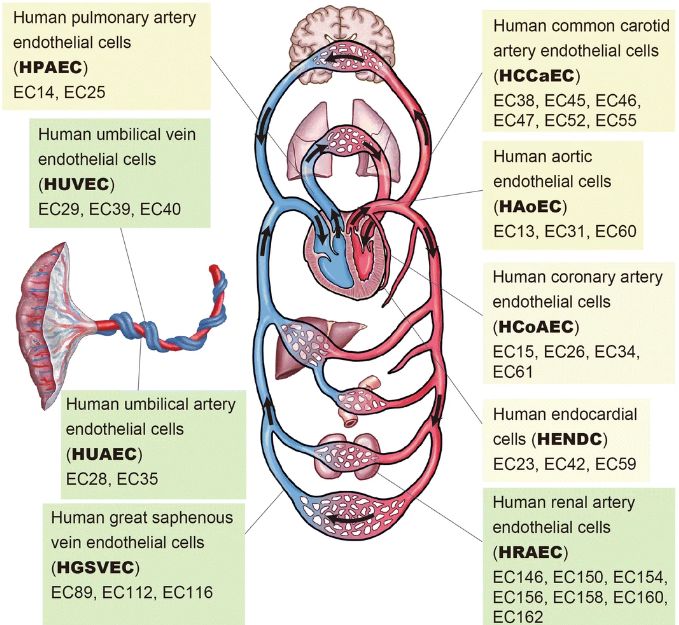
Epigenome and transcriptome database for human vascular endothelial cells
[Nakato et al., Epigenetics & Chromatin, 2019]
